You are what you eat. We’ve all been told this, but the truth is what we eat is often more complex than we are – genetically at least. Take a grain of rice. The plant that produces rice has 40,000 to 50,000 genes, double that of humans, yet we know far more about the composition of the human genome than of plant life. We need to close this knowledge gap quickly if we are to answer the urgent challenge of feeding 8 billion people, especially as food security around the globe is likely to worsen with climate change.
For this reason, AI company InstaDeep has teamed up with Google Cloud to train a large AI model with more than 20 billion parameters on a dataset of reference genomes for cereal crops and edible vegetables, using the latest generation of Google’s Tensor Processing Units (Cloud TPU v4), which is particularly suited for training efficiency at scale. Our aim is to improve food security and sustainable agriculture by creating a tool that can analyze and predict plants’ agronomic traits from genomic sequences. This will help identify which genes make some crops more nutritious, more efficient to grow, and more resilient and resistant to pests, disease and drought.
Genomic language models for sustainable agriculture
Ever since farming began, we have been, directly or indirectly, trying to breed better crops with higher yields, better resilience and, if we’re lucky, better taste too. For thousands of years, this was done by trial and error, growing crops year-on-year while trying to identify and retain only the most beneficial traits as they naturally arise from evolutionary mutations. Now that we have access to the genomic sequences of plants, we hope to directly identify beneficial genes and predict the effect of novel mutations.
However, the complexity of plant genomes often makes it difficult to identify which variants are beneficial. Revolutionary advances in machine learning (ML) can help to understand the link between DNA sequences and molecular phenotypes. This means we now have precise and cost-effective prediction methods to help us close the gap between genetic information and observable traits. These predictions can help identify functional variants and accelerate our understanding of which genes link to which traits – so we can make better crop selections.
Moreover, thanks to the vast library of available crop genetic sequences, training large models on hundreds of plant genomes means we can transfer the knowledge from thoroughly-studied species to those that are less understood but important for food production – especially in developing countries. And by doing this digitally, AI can quickly map and annotate the genomes of both common and rare crop variants.
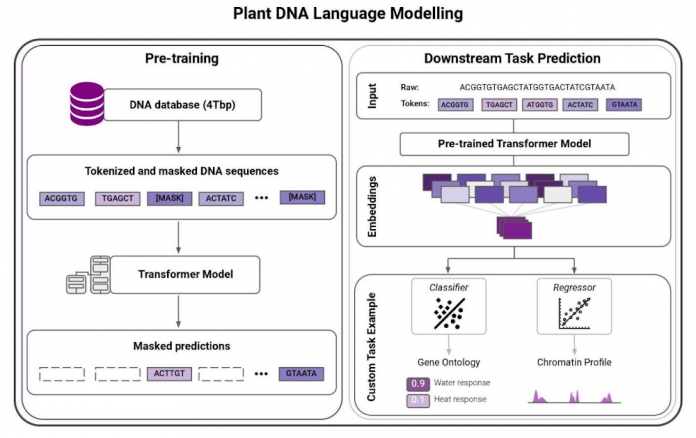
One of the major limitations of traditional ML methods for plant genomics has been they mostly rely on supervised learning techniques. They need labeled data. Such data is scarce and expensive to collect, severely limiting these methods. Recent advances in natural language processing (NLP), such as Transformer architectures and BERT-style training (Bidirectional Encoder Representations from Transformers), allow scientists to train massive language models on raw text data to learn meaningful representations. This unsupervised learning technique changes the game. Once learned, the representations can be leveraged to solve complex regression or classification tasks – even when there is a lack of labeled data.
InstaDeep partners with Google Cloud to train the new generation of AI models for genomics on TPUs
Researchers have demonstrated that large language models can be especially effective in proteomics. To understand how this works, imagine reading amino acids as words and proteins as sentences. The treasure trove of raw genomics data – in sequence form – inspired InstaDeep and Google Cloud to apply similar technologies on nucleotides, this time reading them as words and chunks of genomes as sentences.
Moreover, the representations that the system learned improved in line with the size of the models and datasets, NLP research studies showed. This finding led InstaDeep researchers to train a set of increasingly larger language models on genomics datasets ranging from 1 billion to 20 billion parameters.
Models of 1 billion and 5 billion parameters were trained on a dataset comprising the reference genomes for several edible plants, including fruit, cereal and vegetables for a total of 75 billion nucleotides.
The training dataset must increase in the same proportion as the model capacity, recent work has shown. Thus, we created a larger dataset gathering all reference genomes available on the National Center for Biotechnology Information (NCBI) database including human, animal, non-edible plant and bacteria genomes. This dataset, which we used to train a 20 billion-parameter Transformer model, comprised 700 billion tokens, exceeding the size of most datasets typically used for NLP applications, such as the Common Crawl or Wikipedia dataset.
Both teams announced that the 1 billion-parameter model will be shared with the scientific community to further accelerate plant genomics research.
The compact and meaningful representations of nucleotide sequences learned by these models can be used to tackle molecular phenotype prediction problems. To showcase their ability, we trained a model to predict the gene function and gene ontology (i.e. a gene’s attribute) for different edible plant species.
Early results have demonstrated that this model can predict these characteristics with high accuracy – encouraging us to look deeper at what these models can tell us. Based on these results, we decided to annotate the genomes of three plant species with considerable importance for many developing countries: cassava, sweet potato, and yam. We are working on making these annotations freely available to the scientific community and hope that these will be used to further guide and accelerate new genomic research.
Overcoming scaling challenges with massive models and datasets with Cloud TPUs
The compute requirement for training our 20 billion-parameter model with billions of tokens is massive. While modern accelerators offer impressive peak performance per chip, to utilize this performance often requires tightly coupled hardware and software optimizations. Moreover, maintaining this efficiency when scaling to hundreds of chips presents additional system design challenges. The Cloud TPU’s tightly-coupled hardware and software stack is especially well suited to such challenges. The Cloud TPU software stack is based on the XLA Compiler which offers out-of-the-box optimizations (such as compute and communication overlap) and an easy programming model for expressing parallelism.
We successfully trained our large models for genomics by leveraging Google Tensor Processing Units (TPUv4). Our code is implemented with the JAX framework. JAX provides a functional programming-based approach to express computations as functions that can be easily parallelized using JAX APIs powered by XLA. This helped us to scale from a single host (four chips) configuration to a multi-host configuration without having to tackle any of the system design challenges. The TPU’s cost-effective inter- and intra-communication capabilities led to an almost linear scaling between the number of chips and training time. This allowed us to train the models quickly and efficiently on a grid of 1024 TPUv4 cores (512 chips).
Conclusion
Ultimately, our hope is that the functional characterization of genomic variants predicted by deep learning models will be critical to the next era in agriculture, which will largely depend on genome editing and analysis. We envisage that novel approaches, such as in-silico mutagenesis – the assessment of all possible changes in a genomic region by a computer model – will be invaluable in prioritizing mutations that improve plant fitness and guiding crop improvements. Attempting similar work in wet-lab experiments would be difficult to scale and nearly impossible in nature. By making our current and future annotations available to the research community, we also hope to help democratize breeding technologies so that they can benefit all of global agriculture.
Further Reading
To learn more about the unique features of Cloud TPU v4 hardware and software stack we encourage readers to explore Cloud TPU v4 announcement. To learn more about scaling characteristics, please see this benchmark and finally we recommend reading PJIT Introduction to get started with JAX and SPMD parallelism on Cloud TPU.
This research was made possible thanks to the support of Google’s TPU Research Cloud (TRC) Program which enabled us to use the Cloud TPUv4 chips that were critical to this work.
Cloud BlogRead More