Modeling chemical structures can be a complex and tedious process, even with the help of modern programs and technology. The ability to explore chemical structures at the most fundamental level of atoms and the bonds that connect them is an essential process in drug discovery, pharmaceutical research, and chemical engineering. By infusing chemical research with technology, researchers can expedite outcome timelines, identify hidden relationships, and overall simplify a traditionally complex process.
In this post, we demonstrate how to use Amazon Neptune alongside open-source technologies such as RDKit, and walk through an introduction of modeling chemical structures in a graph database format. We will walk away from this guide being able to ingest, process, visualize, and edit any chemical compound, all within a persistent graph database environment. The following visualization is the molecule caffeine, we will be working towards producing this visualization ourselves throughout this guide.
Solution overview
In order to integrate technology into the analysis of chemical structures, molecules themselves must first be represented in a machine-readable format, such as SMILES (simplified molecular-input line-entry system). SMILES format strings are the industry standard in representing molecular structures. The SMILES format enables the relationships between atoms in a molecular structure to be conveyed as a machine processable string. The SMILES format is not all encompassing, leaving out details such as certain polarities and bond properties. However, SMILES does enable powerful analysis at scale of different structures.
Using Neptune and RDKit, SMILES format data can be ingested, processed, and converted into nodes and edges in a property graph. Modeling molecular structures in a graph database allows for powerful custom visualization and manipulation at the scale demanded by pharmaceutical applications. Utilizing a graph database such as Neptune allows users to compare millions of molecules with millions of associated interactions. Additionally, the fully managed and serverless infrastructure options allow experts with backgrounds in biology and chemistry to focus primarily on the research outcomes of their graph data, avoiding the undifferentiated heavy lifting of managing a complex graph database infrastructure.
This walkthrough follows the process of converting a singular SMILES string, caffeine, to graph data in Neptune. However, the process will work for any SMILES format string you would like to use. We’re sourcing the string for caffeine [CN1C=NC2=C1C(=O)N(C(=O)N2C)C] from the National Library of Medicine, which maintains a public dataset of many chemical structures.
This process also assumes a basic knowledge of Neptune and familiarity with Amazon Neptune graph notebooks. Neptune graph notebooks are Jupyter Notebook interfaces automatically attached to a Neptune database cluster upon creation.
We also use the open-source cheminformatics package RDKit, Python-based data science package Pandas, and AWS SDK for Pandas (awswrangler). RDKit has a strong community and a great number of cheminformatics utilities; we’ll only be exploring a small portion for this post. Pandas is an open-source Python-based data science toolkit with large community support, and AWS SDK for Pandas provides a set of tools to help AWS services interact with Pandas.
There is a copy of the graph notebook utilized in this guide publicly available on the Amazon Neptune github should you want to save time writing code and follow along with a sample notebook in your own environment.
Prerequisites
Please be aware that you will be charged for the AWS resources you use while conducting this walkthrough.
Make sure you have the following prerequisites:
A Neptune cluster.
A Neptune notebook (Python). If you don’t already have a Neptune graph notebook, refer to Use Neptune graph notebooks to get started quickly to create one.
(Optional) Copy of the sample notebook from the Amazon Neptune github
Package setup
The first step in modeling a chemical structure as graph data is importing the required packages. We will be using RDKit, Pandas, and awswrangler
On the Neptune console, navigate to your graph notebook and create a new empty file for Python 3.
Now we need to run the commands to install RDKit, Pandas, and awswrangler within our notebook.
Create a new cell and enter the following code:
Run the cell.
You should receive a response indicating both the download status and the success of the import. If you face any problems even after downloading, restart the kernel for the notebook after the download.
Graph data model
There are a few different options for graph query languages and their associated data models when working with Neptune; in this case we’re using Apache TinkerPop’s Gremlin. We are opting for Gremlin due to its intuitive nature and easy to learn syntax, for those who are new to graph query languages using Gremlin requires us to use a specific data model for our eventual upload of the molecular graph data. For more details on this load format, refer to Gremlin bulk load format.
Create a new cell, enter the following code, then run the cell. This defines the data model for our nodes and edges in the graph. We will be using this newly defined data model to generate data in the required Gremlin formatting for our target compound.
Remember that in this data model, atoms are represented by the nodes, and edges represent the bonds between individual atoms.
RDKit processing
This section is where the chemical computing magic happens. We use the RDKit package to decompose our chemical structure into lists of nodes (atoms) and edges (bonds).
First, we want to declare our SMILES string for the caffeine molecule as a variable. Enter the following code:
To obtain a molecule-type object from RDKit, run the following command in another cell:
Run the following command in another cell to output a 2D picture of our molecule:
The output from the command should look like the following
To recap what we just did, first we declared our SMILES string for caffeine as the variable caffeine_smiles. Next, we used the Chem.MolFromSmiles function from RDKit to turn the SMILES into a Mol type object defined by RDKit. Finally, we returned the Mol type object which resulted in a 2D image of the molecular structure for caffeine that we are working with.
Now we need to iterate through each atom and bond within the mol object outputted from RDKit. While iterating through each atom and bond, we use the graph data model we declared earlier, storing properties of each inside the data model.
Enter the following code into a new cell and run the cell to start the process:
Several different RDKit functions are in this portion of code, so let’s break it down piece by piece:
For the ~id field of both nodes and edges, we combine the data type Node or Edge, the SMILES string itself, and the unique index for the atom
For the ~label field, we use the chemical symbol for nodes, and the bond type for the edges
The fields ~from and ~to for the edges (bonds) are constructed by combining the prefix Node- with the SMILES string, and the respective beginning and ending atoms that the bond connects
The additional fields for the nodes (atoms) in the graph model are the atom’s unique ID within the molecule, its atomic number, and if it is aromatic or not
Note that you can extract several atomic properties for a given SMILES string and add them as additional fields for a given atom or bond. We don’t list them all in this post, but you can explore additional fields for both the atoms and bonds.
Next, we want to transfer our graph data into a Pandas data frame to eventually upload it to the Neptune database.
Enter the following code into a new cell and run the cell:
Now let’s look at all our work so far.
Output a table of the values with the following code in separate cells:
Neptune data load
Now that we have successfully decomposed our caffeine SMILES string into individual atoms and bonds, the next step is to load our data into the Neptune database itself. This will be much simpler than loading data from an external source because our data is already inside the graph notebook environment. This is where the AWS SDK for Pandas comes into play.
First, we need to get the networking details of our Neptune cluster to allow awswrangler to establish a connection to our database. Run the following command to do so.
After running this command, the cell outputs a number of different parameters about your Neptune database configuration. We will focus on only two, the host string and port number.
Next, we need to establish the connection between awswrangler and Neptune, copy the following code into another cell and insert the host and port information from the previous step into the required parameters for the method.
Finally, we will use the .to_property_graph() function of awswrangler to automatically take our Pandas dataframes and insert them as graph data into Amazon Neptune. Copy the following code into another cell and run them to complete to process. Upon success you should receive True as an output from the cell.
You have now successfully uploaded your Pandas dataframes into Amazon Neptune as graph data. From here we can move onto the fun part, visualizing the results of our work so far.
(Optional) Neptune bulk data load
Feel free to skip this section unless you have a large amount of data to upload. If you have a mass amount of data to load into the Neptune storage layer, then you can follow this section to load data in bulk. Loading in bulk will save time compared to the primary method for large data volumes. If you do opt to use this load method be sure to have the proper IAM roles and security configurations enabled in your architecture as outlined in the Neptune documentation. Note, for those following along with the sample notebook from the Neptune github, this section is not included in the notebook file.
First, we need to convert our Pandas data frames to CSV format. Run the following code:
After running this code, we can check our main Jupyter notebook home file system to confirm success.
As shown in the following screenshot, below your Jupyter notebook .ipynb file should be two .csv files labeled caffeine_edges.csv and caffeine_nodes.csv.
Next, we want to load the .csv files we just created into Neptune. To do this, we must download the node and edge files we just created and place them into an S3 bucket.
On the Amazon Simple Storage Service (Amazon S3) console, create a bucket with a custom name of your choice, for example I will use the name: caffeine-molecule-bucket.
Inside this S3 bucket, upload the node and edge files downloaded from Jupyter earlier.
Return to the Neptune workbench.
Run the command %load , to load the .csv files now in Amazon S3. we are using the Neptune workbench magics integrated with the graph notebooks. There are several workbench magics to explore, but we only use %load.
After we run the %load call, we need to complete two fields in order to properly load the data.
For Source, enter the S3 URI for each file in our S3 bucket.
For Load ARN, enter the ARN for an IAM role allowing access for Neptune to read from Amazon S3.
Run the %load cell, first loading the CSV for nodes and then loading the CSV for edges.
You can check on the status of each load in the event of an error by using the line magic %load_status, followed by the load ID output by the previous %load call. Once you have successfully completed the load, the output should look similar to the output below.
Graph network visualization and queries
At this point you have successfully loaded your molecular data into the Neptune database. Now it’s time to visualize the results of our efforts.
First, we can take a look back on the nodes and edges of our graph listed below by running the command nodes_df and edges_df in new cells.
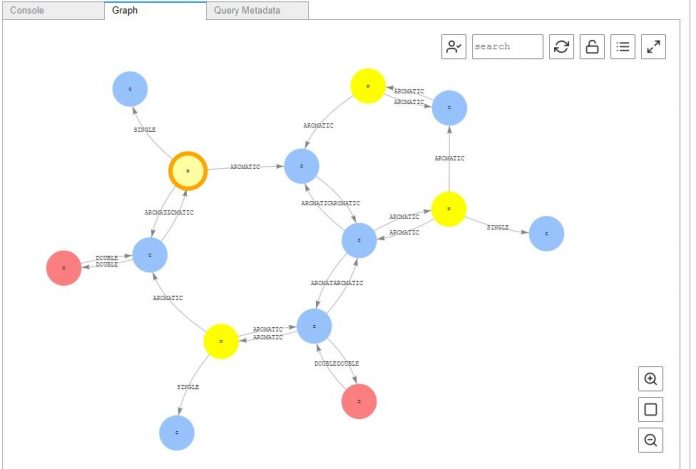
Next, enter the following code into a new cell and run the cell to generate a visualization of your caffeine molecule:
This query is selecting a node in our graph with the label “C” representing the element carbon to start from. Then it is traversing outwards from that node, to a depth of five steps to make sure we cover the whole molecule. Finally, the query records the path the traversal has taken and includes the properties for each item in the graph by adding on the valueMap(true) at the end. While we don’t go into the details of the Gremlin graph query language in this post, if you wish to learn more about the Gremlin language, you can explore its robust documentation.
The visualization should look like the following screenshot.
Explore the nodes and edges further by choosing them individually to see their properties. Now that the data is in our graph database we can digitally explore and save our molecules of choice. We can even add connections to new sections of existing molecules to create and edit our own structures. This can be useful for leaving notes on certain areas of focus on a particular molecule for deeper analysis.
Another option for visualizing your molecule that has been turned into graph data is to utilize the Amazon Neptune Graph Explorer. Graph-explorer is an open-source low-code visual exploration tool for graph data, available under the Apache-2.0 license. It allows you to interact with your molecular graph data without needing to write actual queries. To get started with this React based application simply build the graph-explorer Docker image from the github and deploy it on your compute option of choice.
If you wish to try visualizing additional molecule models you can search for common medical compounds on PubChem. PubChem is a United States government sponsored database of different compounds organized by the National Center for Biotechnology Information. Simply walk back through the beginning of this process and use your SMILES string of choice instead of the caffeine SMILES sting. A few interesting compounds I would suggest are listed below.
Aspirin: “CC(=O)OC1=CC=CC=C1C(=O)O”
Acetaminophen: “CC(=O)NC1=CC=C(C=C1)O”
Azithromycin: “CC[C@@H]1[C@@]([C@@H]([C@H](N(C[C@@H](C[C@@]([C@@H]([C@H]([C@@H]([C@H](C(=O)O1)C)O[C@H]2C[C@@]([C@H]([C@@H](O2)C)O)(C)OC)C)O[C@H]3[C@@H]([C@H](C[C@H](O3)C)N(C)C)O)(C)O)C)C)C)O)(C)O”
Oxytocin: “CC[C@H](C)[C@H]1C(=O)N[C@H](C(=O)N[C@H](C(=O)N[C@@H](CSSC[C@@H](C(=O)N[C@H](C(=O)N1)CC2=CC=C(C=C2)O)N)C(=O)N3CCC[C@H]3C(=O)N[C@@H](CC(C)C)C(=O)NCC(=O)N)CC(=O)N)CCC(=O)N”
Clean up
To avoid any unintended charges to your AWS account, delete your Neptune cluster, Neptune graph notebook, and S3 bucket utilized in this walkthrough when you’re finished.
If you wish to keep your Amazon Neptune resources and associated infrastructure but just clear the data from this walk through out of storage then you can run the following code in another cell.
This will drop the vertices from your nodes dataframe from the database, effectively clearing our graph data. Be sure to also drop any additional nodes you may have added as one-off edits or additions as well.
Conclusion
In this post, you ingested and parsed a SMILES format molecular data string with RDKit and uploaded the individual atoms and bonds as graph data to Neptune. You can replicate this process at scale to accommodate large datasets containing many SMILES strings. You can test this yourself by following the steps for any SMILES string of your choice. With the molecular data broken into individual atoms and bonds in Neptune, you can connect this data to custom bioinformatics applications, chemical computing systems, and research software environments.
You can take this solution even further by integrating Amazon Neptune ML to gain the ability to predict the connections and properties of your molecules.
About the author
Graham Kutchek is a Database Specialist Solutions Architect at AWS with industry expertise in healthcare and life sciences, as well as media and entertainment. He enjoys working with research institutions and healthcare research organizations for the positive impact it has on the world. Connect with him on LinkedIn.
Read MoreAWS Database Blog