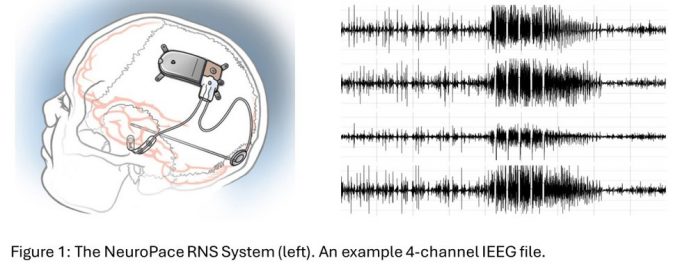
NeuroPace, Inc.1 is a medical device company based in Mountain View, California, that makes the RNS® System,2 a responsive neurostimulation device FDA-approved for treating adults with refractory focal onset epilepsy. The neurostimulator can connect to up to two leads, each with four electrode contacts. It detects patient-specific abnormal patterns programmed by physicians and delivers electrical pulses which are also programmable by physicians. The device captures brain recordings (intracranial EEG or iEEG) which typically hold about 90 seconds of data across four channels, where each channel is sampled at 250 Hz. To date, approximately 16 million iEEG files have been collected from over 5,000 patients.
A key research goal at NeuroPace is to identify effective stimulation patterns for seizure reduction. One hypothesis is that successful treatment settings from previously treated patients with similar iEEG activity may be effective in new patients or in patients who need adjustments to their current stimulation settings, potentially improving upon the conventional trial-and-error approach of finding stimulation programming settings.
For such data-driven approaches to be viable, searching through large iEEG data for similar brain activity patterns must be both scalable and fast, enabling physicians to find similar patient profiles based on selected iEEG files quickly. Previously, finding similar iEEG files across patients required a complex processing pipeline that involved clustering iEEG data within patients, identifying cluster centers, and using dimensionality reduction methods such as PCA and t-SNE to find approximate nearest neighbors (ANN) from other patients. The approximate nearest neighbors were calculated periodically (once every few months) using a limited set of new patient iEEG files, which restricted the tool’s flexibility and practical use.
The good news is recent advancements in vector databases now potentially allow for directly querying the database for similar vector embeddings. This innovation could enable physicians to select any iEEG file from a patient to find similar cross-patient iEEG records without needing to perform clustering steps in advance. The only requirement is to maintain updates to the vector database as new iEEG files become available. This simplification may greatly facilitate querying similar iEEG files across millions of records from thousands of patients, enhancing scalability.
The NeuroPace AI team in collaboration with Google Cloud engineers conducted a proof-of-concept study in which iEEG data was converted into vectors through embedding models and then stored in a vector database – AlloyDB for PostgresQL. AlloyDB is a fully-managed, PostgreSQL-compatible database for demanding transactional workloads, and it supports vector similarity searches based on the pgvector extension. Further, AlloyDB Omni, a downloadable edition of AlloyDB that can be run anywhere, allows the database to be hosted on-premises, keeping the data within the boundaries of an on-prem HIPAA-compliant environment. Additionally, keeping the database on-prem reduces dependence on external network connectivity, which can mitigate the risk of application downtime if the database were hosted externally while the rest of the application remains on-prem.
In this proof-of-concept study, we processed around 1.2 million iEEG files from 414 clinical trial patients.3 Data from 394 patients were added into the AlloyDB cloud service, with the remaining 20 used for testing. Each iEEG file was transformed into a spectrogram image, then into vectors via a custom embedding model developed by the NeuroPace AI team. These vectors were subsequently inserted into the AlloyDB vector database, and 50 randomly selected iEEG files from the test cohort were used to query this database (Figure 2).
Figure 2. Data processing and insertion into AlloyDB Omni
When performing similarity searches, AlloyDB with PGvector has three different index types that can improve latency compared with a brute-force search: Hierarchical Navigable Small World (HNSW), IVFFLAT, and IVF:
HNSW uses a graph-based method, which builds multiple layers of connected nodes for more efficient search pathways, even in large-scale datasets.
The `IVFFLAT` index utilizes a tree-based clustering approach to organize vectors into coarse groups before performing a more detailed search within the most similar clusters; this helps balance both accuracy and search speed.
The new ‘IVF’ index, introduced recently as part of recent AlloyDB AI improvements, leverages Google quantization techniques plus deeper integrations with AlloyDB query processing to both significantly improve query time and increase the total number of dimensions supported per vector.
The fact is that different indexes (and their respective algorithms) can have very different performance with very different use cases. For the NeuroPace use case of finding similar cross-patient iEEGs, we performed extensive benchmarking between IVF and HNSW indexes. Both latency (how quickly the query could complete) and recall (what % of results were present in brute-force queries) were measured. Performance comparisons between these two Approximate Nearest Neighbors (ANN) algorithms indicate that IVF offers high recall rates (~0.9) with a median query latency of about 60 ms, while HNSW indexing resulted in a slightly worse recall rate of 0.8 while also being slower (median query latency of 160 ms) when compared to IVF. Both indexes offer a number of variables to balance between latency and speed.
Nevertheless, both methods significantly outperformed brute force in time to query, with brute force taking nearly 14.7 seconds to find similar iEEG data. A histogram of the query latency and recall for the two different indexing methods compared with brute force are shown in Figure 3. Figure 4 shows the results of the query for one example iEEG query file from a test patient.
Figure 3. Histogram of recall and latency of brute force, and 2 types of indexing IVF and HNSW ANN methods.
Figure 4. An example query iEEG from one of the 20 test patients with electrographic seizure in top 2 channels and a similar iEEG from one of the 394 patients in the search dataset returned by the IVF indexing method.
NeuroPace is enthusiastic about these findings,4 as they hold promise for advancing research in efficiently navigating extensive iEEG data. This advancement has the potential to pave the way for developing algorithms that aid in identifying optimal programming settings for the RNS System. And eventually, we’re excited to try AlloyDB’s new ScaNN index, which may help to further improve performance and usability.
With its lightning-fast vector search built into the reliability of Postgres, all running securely in an off-cloud environment, AlloyDB Omni is facilitating NeuroPace’s research. You can learn more about NeuroPace’s collaboration with Google Cloud, install AlloyDB Omni yourself, or get started with AlloyDB’s managed offering on Google Cloud today! Or get started with vectors on AlloyDB AI with this codelab.
1. https://www.neuropace.com
2. Rx Only. The RNS® System is an adjunctive therapy for adults with refractory, partial onset seizures with no more than 2 epileptogenic foci. See important safety information at http://www.neuropace.com/safety/.
3. Patients consented to the further use of their data for research purposes.
4. The RNS System does not currently incorporate functionality that is based upon or utilizes AI features.
Cloud BlogRead More