DICOM (Digital Imaging and Communications in Medicine) is an image format that contains visualizations of X-Rays and MRIs as well as any associated metadata. DICOM is the standard for medical professionals and healthcare researchers for visualizing and interpreting X-Rays and MRIs. The purpose of this post is to solve two problems:
Visualize and label DICOM images using a custom data labeling workflow on Amazon SageMaker Ground Truth, a fully managed data labeling service supporting built-in or custom data labeling workflows
Develop a DenseNet image classification model using the MONAI framework on Amazon SageMaker, a comprehensive and fully managed data science platform with purpose-built tools to prepare, build, train, and deploy machine learning (ML) models on the cloud
For this post, we use a chest X-Ray DICOM images dataset from the MIMIC Chest X-Ray (MIMIC-CXR) Database, a publicly available database of chest X-Ray images in DICOM format and the associated radiology reports as free text files. To access the files, you must be a registered user and sign the data use agreement.
We label the images through the Ground Truth private workforce. AWS can also provide professional managed workforces with experience labeling medical images.
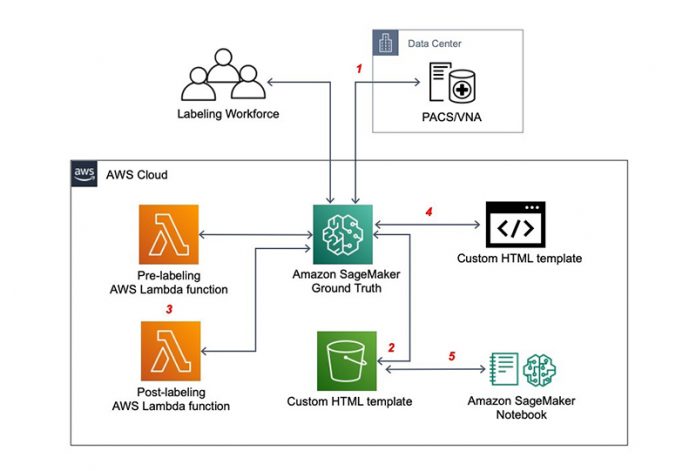
Solution overview
The following diagram shows the high-level workflow with the following key components:
The DICOM images are stored in a third-party picture archiving and communication system (PACS) or Vendor Neutral Archive (VNA), and retrieved through DICOMwebTM.
An input manifest.json file is uploaded to Amazon Simple Storage Service (Amazon S3). The file contains the DICOM instance ID as the data source and potential labels used by annotators when they perform the labeling jobs.
Two AWS Lambda functions are essential for creating labeling jobs on Ground Truth:
A pre-labeling function reads the DICOM image ID from the manifest.json file and creates the task input object that is fed into the Liquid HTML template for automation.
A post-labeling processing function consolidates the annotations from different labeling jobs.
A HTML template with Crowd elements for submitting the labeling jobs and processing the output object. Subsequently, the output of labeling jobs is saved in an output label S3 bucket.
A SageMaker notebook can retrieve the outputs of labeling jobs and use them to train a supervised ML model.
We have built a HTML template that you can use to classify the chest X-Ray DICOM images into one or more categories, out of 13 possible acute and chronic cardiopulmonary conditions. The HTML template built on top of cornerstone.js supports DICOM image retrieval and interactive visualization, plus several annotation and general Cornerstone tools as an example.
In the following sections, we walk through building the DICOM data labeling workflow and performing ML model training using the output of the labeling jobs.
Deploy a third-party PACS on AWS
We use Orthanc as an open-source, lightweight PACS for this post, in which any PACS or VNA supporting DICOMwebTM can be used. You can deploy the Orthanc for Docker container on AWS by launching the following AWS CloudFormation stack:
Fill in the required information during the deployment, including Amazon Elastic Compute Cloud (Amazon EC2) key pair to access the hosting EC2 instance and network infrastructure (VPC and subnets). An NGINX server has been added in the container to proxy the HTTPS traffic to the Orthanc server at port 8042, which also adds Access-Control-Allow-Origin headers for cross-origin resource sharing (CORS). The Orthanc container is deployed on Amazon Elastic Container Service (Amazon ECS) and connected to an Amazon Aurora PostgreSQL database.
After the CloudFormation stack is successfully created, take note of the Orthanc endpoint URL on the Outputs tab.
Create the Lambda functions, S3 bucket, and SageMaker notebook instance
The following CloudFormation stack creates the required Lambda functions with appropriate AWS Identity and Access Management (IAM) roles, S3 bucket for input and output files, and SageMaker notebook instance with sample Jupyter notebook:
For the parameter PreLabelLambdaSourceEndpointURL, enter the Orthanc endpoint URL from previous step, which the pre-labeling task Lambda function uses to generate WADO-URI for a given DICOM instance ID. We recommend creating a notebook instance type of ml.m5.xlarge to carry out the ML modeling after image annotations.
After the stack deployment, take note of the outputs, including the SMGTLabelingExecutionRole and SageMakerAnnotationS3Bucket values.
The IAM role SMGTLabelingExecutionRole is used to create the Ground Truth labeling job. For more information about adding those policies, as well as a step-by-step tutorial on how to run the Ground Truth labeling job after launching the CloudFormation stack, see Build a custom data labeling workflow with Amazon SageMaker Ground Truth.
Upload DICOM images and prepare the input manifest
You can upload the DICOM images to the Orthanc server either through its web UI or the WADO-RS REST API. After the DICOM images are uploaded, you can retrieve the DICOM instance IDs for them, and generate a manifest file with the instance IDs. Each JSON object separated by a standard line break in the manifest file represents an input task sent to the workforce for labeling. The data object in this case contains the instance ID and metadata for the potential labels, which are 13 possible diseases indicated by the image. Assuming one DICOM instance ID is 502b0a4b-5cb43965-7f092716-bd6fe6d6-4f7fc3ce, the corresponding JSON object in the manifest file looks like the following:
After the manifest.json file is compiled, upload it to the S3 bucket created earlier: SageMakerAnnotationS3Bucket.
Build a custom data labeling job:
You should now be able to create a custom labeling job on Ground Truth:
Create a private work team and add members to the team.
The workers receive an email with the labeling portal sign-in URL, which is also available on the Amazon SageMaker console.
AWS can also provide medical imaging experts to label your data. Contact your AWS team for details.
Specify the input and output data locations using the SageMakerAnnotationS3Bucket bucket created earlier.
Specify SMGTLabelingExecutionRole as the IAM role for the labeling job.
For Task category, choose Custom.
Enter the content in liquid.html in the Custom Template text field.
Configure the gt-prelabel-task-lambda and gt-postlabel-task-lambda functions created earlier.
Choose Create.
After you configure the custom labeling task, choose Preview.
The following video shows our preview.
If you created a private workforce, you can go to the Labeling Workforces tab and find the annotation console link.
You can get started with a base HTML template, or modify the sample Ground Truth task UIs for image, text, and audio data labeling jobs. The basic building blocks for the custom template are Crowd HTML elements. The crowd-form and crowd-button are essential for submitting the annotations to Ground Truth. In addition, you need Liquid objects in the template for job automation, in particular, the task input object read in by the pre-labeling Lambda function:
We use the Cornerstone JavaScript library to build the labeling UI that displays a DICOM image in modern web browsers that support the HTML5 canvas element, and use Cornerstone tools to support interactions and enable region of interest (ROI) annotations in different shapes. Additionally, we use the Cornerstone WADO Image Loader to retrieve Web Access to DICOM Objects (WADO) URI through the DICOMwebTM plugin in a remote Orthanc server.
The following two functions are important for retrieving and viewing the DICOM image:
DownloadAndView() – Adds wadouri: to the beginning of the URL so that the cornerstone.js file can find the image loader
loadAndViewImage() – Uses the Cornerstone library to display the image and allows additional functionalities such as zoom, pan, and ROI annotation
You should be able to annotate any DICOM image using the following custom HTML template, which lists the possible labels as multiple option selections for the image classification task.
Build an ML model using the MONAI framework
Now that we have a labeled dataset in the form of the output-manifest.json file, we can start the ML model building process. We created a SageMaker notebook instance earlier through our stack deployment. Alternatively, you can create a new notebook instance. The MONAI library and other additional packages are managed in requirements.txt, which is in the same folder of the training script.
Data loading and preprocessing
After you open the Jupyter notebook, load the output-manifest.json file that has the DICOM URLs and labels.
The following example from dicom_training.ipynb parses the labeled dataset from output-manifest.json:
Now that we have the DICOM URLs in image_url_list and corresponding labels in image_label_list, we can download the DICOM files from Orthanc DICOM server directly into Amazon S3 for SageMaker training:
After you load the images to your S3 bucket, you can display a sample of the images with labels from the parsed JSON file using the MONAI framework:
The following images show a sample output of DICOM images with labels.
We use the MONAI transforms during model training to load the DICOM files directly and preprocess them. MONAI has transforms that support both dictionary and array formats, and is specialized for the high-dimensionality of DICOM medical images. The transforms include several categories such as crop and pad, intensity, I/O, postprocessing, spatial, and utilities. In the following excerpt, the Compose class chains a series of image transforms together and returns a single tensor of the image:
Model training
MONAI includes deep neural networks such as UNet, DenseNet, GAN, and others, and provides sliding window inferences for large medical image volumes. In the DICOM image classification model, we train the MONAI DenseNet model on the DICOM images loaded and transformed using the DataLoader class in the previous steps for 10 epochs while measuring loss. See the following code:
The following example shows how to use the SageMaker Python SDK in a Jupyter notebook to instantiate a training job and run the DICOM model training script:
When you call the fit method in this example, it launches the training instance with a compute size based on the instance_type parameter, instantiates the built-in PyTorch deep learning container, installs the MONAI dependent packages found in requirements.txt, and runs the training script referenced by the entry_point=’monai_dicom.py’ parameter.
For more details on how to use the MONAI framework within SageMaker to build and deploy your medical image models, see Build a medical image analysis pipeline on Amazon SageMaker using the MONAI framework. To learn about other custom data labeling workflows, see Build a custom data labeling workflow with Amazon SageMaker Ground Truth.
Conclusion
This post showed how to build a DICOM custom labeling workflow with Ground Truth. Additionally, we showed you how to take the output manifest file generated from your labeling workflow and use it as a labeled dataset to build a DenseNet image classification model using the MONAI framework.
If you have any comments or questions about this post, please use the comments section. To find the latest developments to this solution, check out the GitHub repo.
About the Authors
Nihir Chadderwala is an AI/ML Solutions Architect on the Global Healthcare and Life Sciences team. His background is building big data and AI-powered solutions to customer problems in a variety of domains such as software, media, automotive, and healthcare. In his spare time, he enjoys playing tennis, and watching and reading about Cosmos.
Gang Fu is a Senior Healthcare Solutions Architect at AWS. He holds a PhD in Pharmaceutical Science from the University of Mississippi and has over 10 years of technology and biomedical research experience. He is passionate about technology and the impact it can make on healthcare.
Bryan Marsh is a Senior Solutions Architect in the Academic Medical Center team at Amazon Web Services. He has expertise in enterprise architecture with a focus in the healthcare domain. He is passionate about using technology to improve the healthcare experience and patient outcomes.
Read MoreAWS Machine Learning Blog